Projects
News: Video to present Traumatrix. Statitical challenges slides. Trauma conference london 2024, slides. NewsLetter TraumaNews May 2023.
Old news: Video 2019 of presentation of the project and collaboration with Capgemini Invent –Short slides presentation 2019. -Medias: Podcast Pharmaradio – revue de presse – article BioWorldMedTech
For nearly eight years, I have been collaborating with a team of intensive care clinicians specializing in trauma patients—those involved in road accidents, falls, or similar incidents, often suffering from head injuries or hemorrhagic shocks. Our group operates as a public/private consortium involving École des hautes études en sciences sociales, École Polytechnique, Inria, CNRS, and Capgemini Invent, supported through a skills sponsorship. Our work has focused on improving data collection (data from the accident scene to hospital discharge), developing causal inference methods to assess treatment effectiveness, recommending therapeutic strategies (e.g., optimal transfusion doses, personalized treatments, and timing), managing missing information, and creating predictive models to aid decision-making and patient management in high-stakes, time-sensitive environments.
We are now starting real-time model evaluation in collaboration with SAMU, with plans to deploy predictive models in French ambulances to measure their impact on patient care. The project presents numerous scientific challenges: ensuring predictive models are robust to changes in practices and patient profiles, evaluating algorithmic fairness (e.g., equitable performance across genders), building confidence in predictions (e.g., algorithms acknowledging uncertainty with « I don’t know »), designing effective human-machine interactions to present results to doctors, and integrating heterogeneous data from European counterparts. Bridging these gaps is essential to successfully translate innovations into clinical practice.
The Traumatrix project presents numerous benefits for academic research and is a source of motivation for doctoral and postdoctoral researchers who can quickly test innovations against real-world data and contribute to a societal project. In addition, transitioning from method development to real-time implementation is a particularly exciting.

Respiratory Diseases:
I work on respiratory diseases with colleagues from Inserm, such as asthma. Even though these diseases result from genetic-environmental interactions, the WHO estimates that by 2050, one in two people will suffer from respiratory diseases, allergies, asthma, etc., due to climate change, air quality, biodiversity loss, etc. This is a major public health issue. To improve prevention, establish new diagnostics and therapeutic practices, we use an exposomic approach. This means we study all social and environmental factors that, combined with patient characteristics, help predict the onset and progression of these diseases. AI is useful for accelerating research by using heterogeneous, multidimensional, and complex data. This is all the more important as the randomised control trial population only covers 10% of those who will receive treatment (Pahus, et. al. 2019), so there is a real need to provide more evidence to know how best to treat patients.
ICUBAM: ICU Bed Availability Monitoring
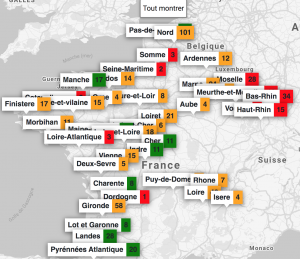
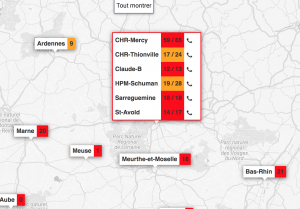
One of the major challenges during the health crisis was the availability of resuscitation beds equipped with ventilators. To deal with this problem, we created with colleagues from Inria and a team of computer scientists, clinicians and researchers, ICUBAM as an operational tool for resuscitators to monitor and visualize bed availability in real time. Resuscitators directly provided data on available beds as well as demographic data on mortality, entries and transfers on their mobile phone (in less than 10 seconds) and got an updated visualization on the map of France.
The project described in an article, in an interview and in slides, is the result of a personal initiative of a resuscitator (A. Kimmoun) from the Grand Est region severely affected by the crisis.
In less than two weeks, all intensive care units in this region were overwhelmed and beds were created spontaneously. Official information systems were unable to provide reliable and up-to-date information on bed availability and this information was crucial for efficient allocation of patients and resources. Due to my collaborations with a network of resuscitators, I organized a meeting on March 22, 2020 with colleagues Gabriel Dulac Arnold, Olivier Teboule and A. Kimmoun and we worked 7 days a week for several weeks on the project building an inter-disciplinary team (Laurent Bonnasse-Gahot EHESS, Maxime Dénès Inria, Sertan Girgin Google Research, François Husson CNRS, IRMAR, Valentin Iovene Inria, François Landes Université Paris-Saclay, Jean-Pierre Nadal CNRS & EHESS, Romain Primet Inria, Frederico Quintao Google Research, Pierre Guillaume Raverdy Inria, Vincent Rouvreau Inria, Roman Yurchak). ICUBAM was deployed on March 25 with the agreement of the ARS (regional health agency) EST and was used by 40 departments, 130 resuscitation services and covered more than 2,000 resuscitation beds.
The success of ICUBAM can be explained on the one hand by the quality of the data collected (directly at the bedside) which allows a real-time inventory and a modelling of the evolution of the epidemic to inform patients, health authorities and practitioners and on the other hand by a team with complementary skills working with caregivers and institutions.
Distributed matrix completion for medical databases
This started as a joint work with Geneviève Robin (CNRS Researcher), François Husson (Professor at Agrocampus Ouest) and Balasubramanian Narasimhan (Senior Researcher at Stanford University) and Anqi Fu (Phd Student at Stanford). Personalized medical care relies on comparing new patients profiles to existing medical records, in order to predict patients treatment response or risk of disease based on their individual characteristics, and adapt medical decisions accordingly. The chances of finding profiles similar to new patients, and therefore of providing them better treatment, increase with the number of individuals in the database. For this reason, gathering the information contained in the databases of several hospitals promises better care for every patient. However, there are technical and social barriers to the aggregation of medical data. The size of combined databases often makes computations and storage intractable, while institutions are usually reluctant to share their data due to privacy concerns and proprietary attitudes. Both obstacles can be overcome by turning to distributed computations, which consists in leaving the data on sites and distributing the calculations, so that hospitals only share some intermediate results instead of the raw data. This could solve the privacy problem and reduce the cost of calculations by splitting one large problem into several smaller ones. The general project is described in Narasimhan et. al. (2017). As it is often the case, the medical databases are incomplete. One aim of the project is to impute the data of one hospital using the data of the other hospitals. This could also be an incentive to encourage the hospitals to participate in the project and to share their summaries of their data. This project is continued with Claire Boyer, Aurélien Bellet, Marco Cuturi.
Missing Values: more ressources on Rmistatic
The problematic of missing values is ubiquitous in data analysis. The naive workaround which consists in deleting observations with missing entries is not an alternative in high dimension as it will lead to the deletion of almost all data and huge biais. The methods available to handle missing values depend on the aim of the analysis, the pattern of missing values and the mechanism that generates missing values. Rubin in 1976 defined a widely used nomenclature for missing values mechanisms: missing completely at random (MCAR) where the missingness is independent of the data, missing at random (MAR) where the probability of being missing depends only on observed values and missing not at random (MNAR) when the probability of missingness then depends on the unobserved values. Large part of the literature focuses on MCAR and MAR. In Näf, J. Scornet, E. & Josse, J. (2024) What Is a Good Imputation Under MAR Missingness?, we clarified what is MAR and provide a typology of mechanism using the pattern mixture point of view.
Contribution on MNAR data (identifiability and estimation):
- Sportisse, A., Boyer, C. & Josse, J. (2020). Estimation and Imputation in Probabilistic Principal Component Analysis with Missing Not At Random Data. NeurIPS.
- Sportisse, A., Boyer, C. & Josse, J. (2020) Imputation and low-rank estimation with Missing Not At Random data. Statistics & Computing.
Many statistical methods have been developed to handle missing values (Little and Rubin, 2019; van Buuren, 2018) in an inferential framework, i.e. when the aim is to estimate parameters and their variance from incomplete data. One popular approach to handle missing values is imputation, which consists in replacing the missing values by plausible values to get a completed data that can be analyzed by any methods. Mean imputation is the worst thing that can be done in an inferential framework as it distorts joint and marginal distribution. One can either impute according to a joint model or using a fully conditional modelling approach. Powerful methods include imputation by random forest (using misforest package) but also imputation by low rank methods (using missMDA, softimpute packages) and recently using optimal transport.
Contribution on imputation methods (with SVD based methods, optimal transport):
- Näf, J., Scornet, E. & Josse, J. (2024). What Is a Good Imputation Under MAR Missingness?
- Muzellec, B., Josse, J. Boyer, C. & Cuturi, M. (2020) Missing Data Imputation using Optimal Transport. ICML2020.
- Mozharovskyi, P., Josse, J. , Husson, F. (2020). Nonparametric imputation by data depth. Journal of the American Statistical Association.
- Husson F., Josse J., Narasimhan B., Robin, G. (2019) Imputation of mixed data with multilevel singular value decomposition. Journal of Computational and Graphical Statistics.
- Audigier, V. Husson, F. Josse, J. (2016). A principal components method to impute missing values for mixed data. Advances in Data Analysis and Classification.
A single imputation method can be interesting in itself if the aim is to predict as well as possible the missing values (to do matrix completion). Nevertheless, even if we manage to impute by preserving as well as possible the joint and marginal distribution of the data, a single imputation can not reflect the uncertainty associated to the prediction of missing values. To achieve this goal, multiple imputation (MI) (van Buuren, 2018 in the mice package) consists in generating several plausible values for each missing data (to reflect the variance of prediction given observed data and imputation model) leading to different imputed data sets. Then, the analysis is performed on each imputed data sets and results are combined so that the final variance takes into account the supplement variability due to missing values.
Contribution on multiple imputation methods (based on low rank methods):
- Audigier, V. Husson & F. Josse, J. (2017). MIMCA: Multiple imputation for categorical variables with multiple correspondence analysis. Statistics and Computing.
- Audigier, V., Husson, F. & Josse, J. (2015). Multiple Imputation with Bayesian PCA. Journal of Statistical Computation and Simulation.
- Josse, J., Husson, F. & Pagès, J. (2011). Multiple imputation in PCA. Advances in data analysis and classification.
An alternative to handle missing values consists in modifying estimation processes so that they can be applied to incomplete data. For example, one can use the EM algorithm to obtain the maximum likelihood estimate (MLE) despite missing values. This is implemented for instance for regression and logistic regression in the R package misaem.
Contribution on methods to do inference with missing values (logistic regression, variable selection):
- M. Bogdan, W. Jiang, J. Josse, B. Miasojedow & V. Rockova. (2021). Adaptive Bayesian SLOPE – High dimensional Model Selection with Missing Values. JCGS.
- Jiang, W., Lavielle, M. Josse, J. & T. Gauss. (2019). Logistic Regression with Missing Covariates — Parameter Estimation, Model Selection and Prediction within a Joint-Modeling Framework. CSDA
Contribution on methods to do exploratory data analysis with missing values; combine estimation and imputation (PCA with missing values):
- How to perform a PCA with missing values? How to impute data with a PCA?. François Husson Youtube playlist
- Josse, J & Husson, F. (2015). missMDA a package to handle missing values in and with multivariate data analysis methods. Journal of Statistical Software.
- Josse, J & Husson, F. (2013). Handling missing values in exploratory multivariate data analysis methods.
- Husson, F. & Josse, J. (2011). Handling missing values in Multiple Factor Analysis. Food Quality and Preferences.
Finally, for supervised learning with missing values, where the aim is to predict as well as possible an outcome and not to estimate parameters as accurately as possible, the solutions are very different. With our group, we have suggested new approaches to tackle this issue. For instance, we show that the solution which consists in imputing the train and the test set with the means of the variables in the train set, even if this is not appropriate for estimation is consistent for prediction. We have also studied solution to do random forest with missing values, which consists in using the missing incorporated in attributes criterion (implemented in scikitlearn in HistGradientBoosting) and in the R grf package. Finally, we have developed new methods theoretically justified to do neural nets with missing entries.
Contribution on supervised learning with missing values (SGD, random forest, neural nets):
- Josse, J., Jacob, M. Chen, Prost, N., Varoquaux, G Scornet, E. On the consistency of supervised learning with missing values (2019-2024). Statistical Papers.
- Le Morvan, J. Josse, E. Scornet. & G. Varoquaux (2021). What’s a good imputation to predict with missing values?
Neurips 2021. (Spotlight). - A. Sportisse, C. Boyer, A. Dieuleveut, J. Josse (2020). Debiasing Stochastic Gradient Descent to handle missing values. Neurips2020.
- Le Morvan, J. Josse, M., Moreaux, T, E. Scornet. & G. Varoquaux (2020). Neumiss networks: differential programming for supervised learning with missing values. Neurips2020. (Oral)
- Le Morvan, M., N. Prost, J. Josse, E. Scornet. & G. Varoquaux (2020). Linear predictor on linearly-generated data with missing values: non consistency and solutions. AISTAT2020.